Clinical & Medical Surgery
Open Access
Volume 3
Ju-Hong Yang1; Hai-Ying Jiang2; Song Zhang2*
*Corresponding Author: Song Zhang
Department of Otolaryngology, University of Chinese Academy of Science-Shenzhen Hospital, Song Bai Road 4253, Shenzhen, Guang Dong TX 518106, PR China.
Email: zhangsongent@foxmail.com
Article Info
Received: Jan 07, 2023
Accepted: Feb 03, 2023
Published: Feb 10, 2023
Archived: www.jclinmedsurgery.com
Copyright: © Zhang S (2023).
Abstract...
Objectives: This study aimed to discuss the role of the feline sarcoma oncogene (FES) in the occurrence and development of nasopharyngeal carcinoma. Methods: Fluorescence quantitative polymerase chain reaction (PCR) and western blotting were applied to the detection of the FES gene and protein expression in tissue samples of 10 nasopharyngeal carcinoma cases and 10 chronic nasopharyngitis cases. Results: The FES gene and protein expression level in chronic nasopharyngitis was higher than in nasopharyngeal carcinoma, with statistically significant difference (P<0.05). Conclusions: High expression level of the FES gene and protein can suppress the occurrence and development of nasopharyngeal carcinoma.Citation: Yang JH, Jiang HY, Zhang S. The Role of FES Gene Expression in the Occurrence of Nasopharyngeal Carcinoma. J Clin Med Surgery. 2023; 3(1): 1073.
Background
The mechanism of occurrence and development of nasopharyngeal carcinoma is not quite clear at present, and it may be related to genetic inheritance, geographic environment, living habits, infection by the Epstein-Barr virus and many other factors. Recent years have seen increasing attention to studies on some carcinoma-associated genes, such as activation of protooncogenes and deactivation anti-oncogenes, both of which will lead to unlimited proliferation of cells and ultimately to carcinomas. Genetic changes, such as mutation and deletion, can modify the expression of a gene and therefore influence its function. This is especially true with proto-oncogenes and anti-oncogenes, the changes of which may affect the proliferability of cells, thereby initiating tumor or suppressing tumor growth. In this study, the authors discussed the role the Feline Sarcoma Oncogene (FES) plays in the occurrence and development of nasopharyngeal carcinoma by detecting and comparing the expression of FES in nasopharyngeal carcinoma and chronic nasopharyngitis tissues.
Data and methods
Clinical Data
The samples used in this study included 10 nasopharyngeal carcinoma and 10 chronic nasopharyngitis tissue samples, all coming from patients suspected of having nasopharyngeal neoplasm and treated by our hospital. The patients had not undergone radiotherapy or chemotherapy before biopsy. The 10 nasopharyngeal carcinoma patients, 7 males and 3 females, consist of 2 T1 cases, 4 T2 cases, 3 T3 cases and 1 T4 case, according to the 8th edition of the UICC TNM Classification. Six cases presented with lymph node metastasis, while the other 4 had no such metastasis.
Fluorescence quantitative Polymerase Chain Reaction (PCR)
TRIzol reagent was used to extract the total RNA of the above tissues. 1 μl of RNA was obtained from 3 randomly selected samples for gel electrophoresis (1% agarose gel, 80 V x 20 min), a gel imaging system was used to observe the 5s rRNA, 18s rRNA and 28s rRNA bands of the total RNA, and the completeness of all three bands indicated the comparatively complete extraction of total RNA. Reverse transcription was performed with reverse transcriptase to form first-strand (cDNA). FES and β-actin primers were designed respectively based on the gene sequences in the GeneBank and put under PCR with cDNA as the template. FES primers: 5’-TGTTGGGTGAGCAGATTGGA-3’(S), 5’-TGGGAGCGTCTCTCGACAA-3’(R); β-actin primers: 5’- GCATGGGTCAGAAGGATTCCT-3’(S), 5’-TCGTCCCAGTTGGTGACGAT-3’. Reaction system: H2O 6.5 μl, Premix EX TaqTM (Probe qPCR) 2 × 10 μl, forward primer (10 pmol/μl) 0.5 μl, reverse primer (10 pmol/μl) 0.5 μl, cDNA 2 μl, making a total volume of 20 μl. The annealing temperature was 60oC for all samples. The relative expression of samples=2-△△CT.
Western blot
The total protein contained in the above tissues was extracted with the total protein extraction kit (Applygen Technologies Inc., Beijing, China) following the manual, and was quantified with the BCA protein quantitation kit. The protein western blotting was conducted in the following steps: making SDS-PAGE gel, protein loading, electrophoresis, transfer onto a membrane, blocking, primary antibody incubation, secondary antibody incubation, visualization, taking pictures with a digital gel image analysis system, analyzing optical density and recording results.
Statistical analysis: SPSS18.0 software was used for data collection and analysis, and independent samples t test was applied to analyze the difference in the relative expression levels of FES mRNA and grey values of the FES protein bands, with the difference being significant when P<0.05.
Results
Fluorescence quantitative PCR
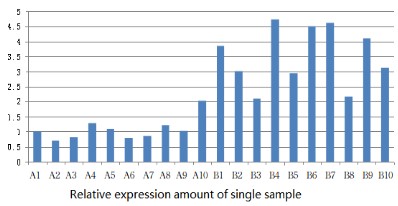
Fluorescence quantitative PCR technique detected the expression of FES mRNA and the results were represented in the relative expression per single sample (Figure 1). The relative expression of FES mRNA in chronic nasopharyngitis tissues (3.52 ± 0.98) was significantly higher than in nasopharyngeal carcinoma (1.08 ± 0.38), t=7.29, and P<0.05, so the difference was statistically significant (Table 1).
Western blot
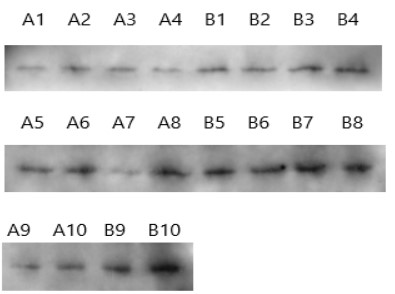
Western blotting was used to detect the protein expression in chronic nasopharyngitis and nasopharyngeal carcinoma (Figure 2). The average optical density of protein bands of the nasopharyngeal carcinoma group (433.67 ± 162.61) was significantly lower than that of the chronic nasopharyngitis group (704.91 ± 170.90), and t=3.64, P<0.05, so the difference was statistically significant (Table 2).
Table 1: Relative expression amount of FES gene in a single sample.
| Group | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 1.0 | 0.70 | 0.81 | 1.27 | 1.10 | 0.80 | 0.86 | 1.22 | 1.04 | 2.04 |
| B | 3.87 | 3.02 | 2.10 | 4.75 | 2.93 | 4.51 | 4.62 | 2.17 | 4.11 | 3.14 |
A: Nasopharyngeal carcinoma tissue; B: Chronic nasopharyngitis tissue. t=3.64; P<0.05.
Table 2: Average optical density of FES protein bands.
| Group | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 254.1 | 400.2 | 351.9 | 260.1 | 535.8 | 680.3 | 321.1 | 720.0 | 391.5 | 421.6 |
| B | 596.3 | 377.1 | 607.2 | 634.7 | 766.3 | 604.1 | 780.5 | 870.0 | 883.6 | 929.3 |
A: Nasopharyngeal carcinoma tissue; B: Chronic nasopharyngitis tissue. t=7.29; P<0.05.
Discussions
Without clear pathogenesis, nasopharyngeal carcinoma is a common malignant tumor in the ENT department. Despite a comparatively large number of studies on the expression of relevant oncogenes or anti-oncogenes in recent years, the pathogenic genes of nasopharyngeal carcinoma are still undefined. In this study, the authors detected the expression levels of the FES gene and protein in 10 nasopharyngeal carcinoma and 10 chronic nasopharyngitis tissue samples. Fluorescence quantitative PCR was used to detect the relative expression per single sample, with results demonstrating higher expression of the FES gene in chronic nasopharyngitis tissue than in nasopharyngeal carcinoma tissue. Western blot analysis also revealed higher expression level of the FES protein in chronic nasopharyngitis tissue. These results indicated that the FES gene might suppress the occurrence and development of nasopharyngeal carcinoma, and it might be an anti-oncogene.
A member of the subfamily of the FES-associated non-receptor protein tyrosine kinases [1], the FES protein has the activity of tyrosine-specific protein kinases, and contains a central Src homology domain and a tyrosine kinase catalytic domain at the C-terminal end [2]. FES was initially identified as a retroviral oncogene that causes feline carcinomas [3]. Recently, Sangrar [4] discovered that FES had obvious cancer suppressing function in breast cancer, Kuo’s [5] research revealed silenced expression of the FES gene due to methylation in esophageal cancer, Christensen [6] pointed out that such silenced expression might be related to the grading of breast cancer, and Yang et al. [7] found that overexpression of FES could suppress the invasion and metastasis of osteosarcoma cells. However, some studies revealed that abnormally inactivated FES might have something to do with the proliferation, metastasis and invasion of several carcinomas [8,9], and downregulation of FES expression might suppress the proliferation of tumor cells. The opposite functions FES performs in different carcinomas can be attributable to its dual identity as both a proto-oncogene and an anti-oncogene, which enables it to play different roles in different tissues.
Conclusion
In summary, our test results showed that the high expression of FES could suppress the occurrence and development of nasopharyngeal carcinoma, which meant that FES acted as a tumor suppressor in nasopharyngeal carcinoma tissues.
Declarations
Ethics approval and consent to participate: Ethics committee of University of Chinese Academy of Science-Shenzhen Hospital approved the study. The reference numberis 20221109.
Conflict of interest: The authors report no conflict of interest.
Funding: This work was supported by the Guangming District Economic Development Special Fund (2020R01078, 2021R01064).
Authors' contributions: Song Zhang designed the study and supervised all experiments. Ju-HongYang executed RT-PCR and Western Blot experiments, and drafted this paper. Hai-Ying Jiang executed data sorting. All authors read and approved the final manuscript.
References
- Smithgall TE, Rogers JA, Peters KL, Li J, Briggs SD, et al. The c-Fes family of protein-tyrosine kinases. Crit Rev Oncog. 1998; 9: 43.
- Hjermstad SJ, Peters KL, Briggs SD, Glazer RI, Smithgall TE. Regulation of the human c-fes protein tyrosine kinase (p93c-fes) by its src homology 2 domain and major autophosphorylation site (Tyr-713). Oncogene. 1993; 8: 2283.
- Shibuya M, Hanafusa T, Hanafusa H, Stephenson JR. Homology exists among the transforming sequences of avian and feline sarcoma viruses. Proc Natl Acad Sci USA. 1980; 77: 6536-6540.
- Sangrar W, Zirgnibl RA, Gao Y, Muller WJ, Jia Z, et al. An identity crisis for fps/fes: oncogene or tumor suppressor? Cancer Res. 2005; 65: 3518-3522.
- Kuo I-Y, Chang J-M, Jiang S-S, Chen C-H, Chang I-S, et al. Prognostic CpG methylation biomarkers identifed by methylation array in esophageal squamous cell carcinoma patients. Int J Med Sci. 2014; 11: 779.
- Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, et al. Breast cancer DNA methylation profles are associated with tumor size and alcohol and folate intake. PLoS Genet. 2010; 6: e1001043.
- Zhao Y, Wang Z, Wang Q, Sun L, Li M, et al. Overexpression of FES might inhibit cell proliferation, migration, and invasion of osteosarcoma cells. Cancer Cell International. 2020 ; 20: 102.
- Voisset E, Lopez S, Dubreuil P, De SP. The tyrosine kinase FES is an essential efector of KITD816V proliferation signal. Blood. 2007; 110: 2593.
- Shigeru K, Yasuyoshi M, Hiroshi K, Smithgall TE. Downregulation of the c-Fes protein-tyrosine kinase inhibits the proliferation of human renal carcinoma cells. Int J Oncol. 2009; 34: 89.